What are metagenomics?Metagenomics also known as environmental genomics, ecogenomics, or community genomics is the study of metagenomes. Metagenomes are genetic material directly recovered from environmental samples. It's a newer field in which genomic analysis is applied to entire microbial communities. A better understanding of how microbial communities survive with pollution can be used to better strategize monitoring the impact of pollution on the ecosystem, and for cleaning up contaminated environments. Originally the term “Metagenomics” coined by Jo Handelsman and others in University of Wisconsin Department of Plant Pathology, in 1998, referenced the idea that a collection of genes sequenced from the environment could be analyzed in a way similar to the study of a single genome.  However recently researchers Kevin Chen and Lior Pachter of the University of California, Berkley defined metagenomics as “the application of modern genomics techniques to study of communities of microbial organisms directly in their environments, bypassing the need for isolation and lab cultivation of individual species.” Metagenomics is an extremely exciting endeavor, because the tools of classic genomics and microbiology largely rely on isolating individual microbial species in pure cultures, cultures containing only microbes of a particular species. This means that there is still a vast microbial world out there that has been inaccessible to science. Scientist estimate less than 1% of the estimated millions of microbial species on Earth can be cultured. By allowing scientists to access a community's genome without relying on pure cultures, metagenomics transcend the limitations of classic genomics and microbiology. Most microbial species exist at low amounts, they resist cultivation, and have escaped detection by DNA-based analyses. These organisms are likely factors to community stability and function, and represent an enormous pool of uncharacterized genes and enzymes, useful for medical and industrial purposes. Metagenomics is a great proses to cultivate all the unknown microorganisms. |
 Yet there are some challenges that scientists face. For example scientists must determine how to best sample an environment, how many times samples should be taken, and wether a sample is representative of the environment. Improved DNA extracted from a sample adequately represents the entire community’s genome and has little or no contamination. Scientists need databases that use common standards and are accessible to the people. It is a field that needs more institutional frameworks, a broad range of fields; like chemistry, genetics, microbiology, biochemistry, pathology, ecology, evolution, soil and atmospheric sciences, geology, oceanography, statistics, computer sciences, database development, mathematics, engineering, and others have applicability to metagenomics.
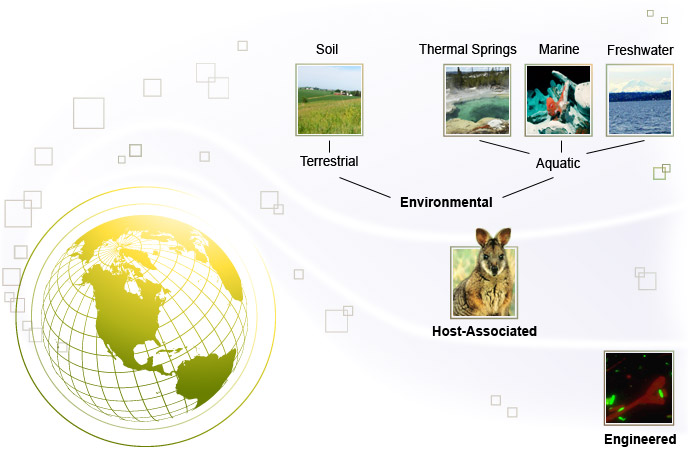
Today metagenomics has been used to analyze different microbial communities such as soil, oral, fecal, and aquatic ecosystems.
A great example of a practical application of metagenomics can be seen in the video below. |